Exploring the potential of machine learning to empower image-guided and robot-assisted interventions.
MRI became a crucial diagnostic tool for prostate cancer care. Recent progress in multiparametric MRI (mpMRI) and the PI-RADS reporting system greatly improved radiologists’s ability to detect, evaluate, and report potential diseases. However, raw MRI data, along with a conventional free-text report, do not fully convey the radiologists’ findings, such as the tumor location and its proximity to critical structures, and thus are not a suitable format for the planning and navigation of surgical interventions (e.g., biopsy, ablations, radiation, and surgery), especially when those tasks are computerized. They must be segmented and labeled to create a patient-specific geometric model.
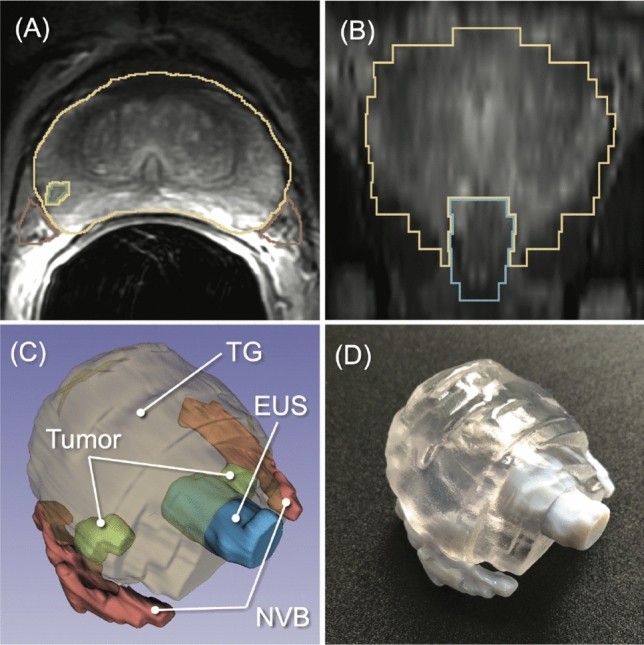
To streamline the process of creating a patient-specific geometric model from prostate MRI data, we investigated a new machine-learning algorithm to automatically segment the prostate and surrounding critical structures, including the neurovascular bundles (NVBs) and the external urethral sphincter (EUS), on the images. We used labeled MRI data from our previous clinical study to train a convolutional neural network (CNN) model based on 3D U-Net.
The specific challenge in this effort was that the label data for those structures were not widely available and were created only from MRI data obtained with a single imaging protocol at a single site. As network models trained on data from a single source suffer from quality loss due to the domain shift, we propose a semi-supervised domain adaptation (DA) method to refine the model’s performance in the target domain. Our DA method combines transfer learning and uncertainty-guided self-learning based on deep ensembles. Experiments on the segmentation of the prostate, NVB, and EUS, show significant performance gains with the combination of those techniques compared to pure TL and the combination of TL with simple self-learning.
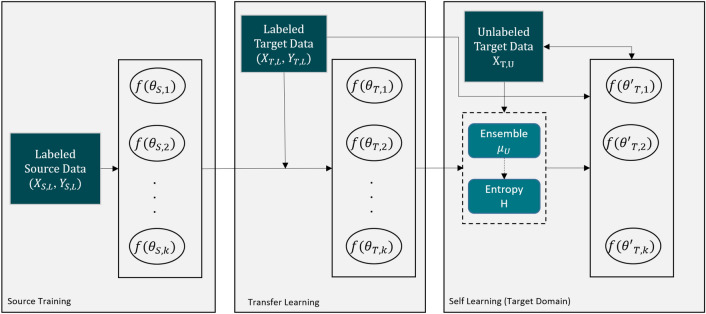
Results on a different task and data demonstrate our method’s generic application capabilities. Our method has the advantage that it does not require any further data from the source domain, unlike the majority of recent domain adaptation strategies. This makes our method suitable for clinical applications where the sharing of patient data is restricted.
References
- Meyer A, Mehrtash A, Rak M, Bashkanov O, Langbein B, Ziaei A, Kibel AS, Tempany CM, Hansen C, Tokuda J. Domain adaptation for segmentation of critical structures for prostate cancer therapy. Sci Rep. 2021;11(1):11480. doi:10.1038/s41598-021-90294-4 PMID: 34075061. PMCID: PMC8169882.
Acknowledgements
The study was funded in part by the U.S. National Institutes of Health (R01EB020667, R01CA235134, P41EB015898, P41EB028741), and the EU and the federal state of Saxony-Anhalt, Germany (ZS/2016/08/80388).


